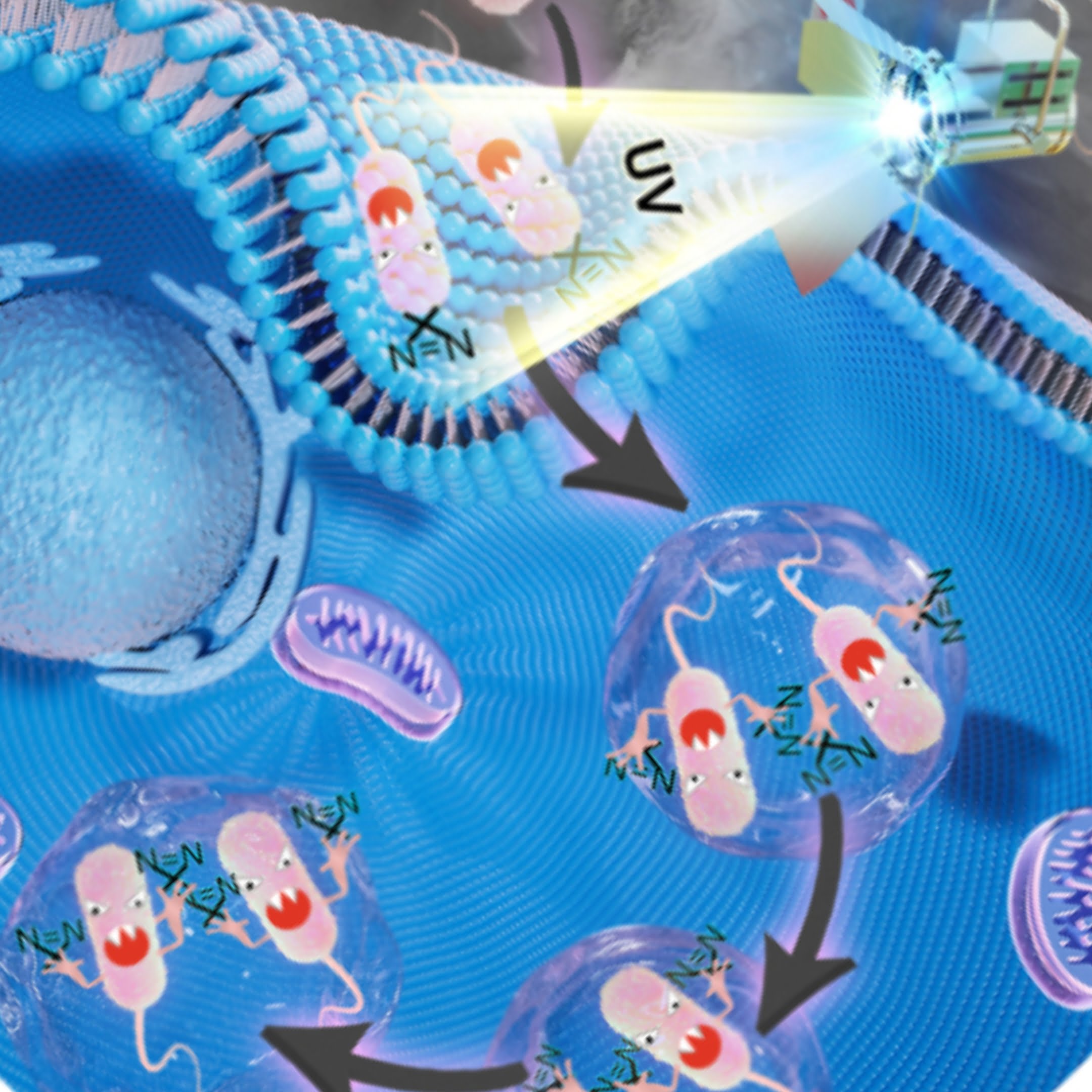
When bacteria like salmonella infect and sicken people, they hijack a person’s cell proteins to develop a defense against an immune response. Understanding how that works and developing methods for defending against these bacteria is difficult because scientists haven’t been able to track the hundreds of proteins involved in real time.
Now, W. Andy Tao, a Purdue University professor of biochemistry, and colleagues at Purdue and Fudan University in China, have developed a chemical method — host and pathogen temporal interaction profiling, or HAPTIP — for labeling a living bacteria and tracking it as it invades a host cell. Their findings, published in the journal Angewandte Chemie, may help improve understanding of bacterial infections and lead to the development of new drugs.
“The interaction between host cells and pathogens are highly dynamic and complex with many questions to be answered. It is extremely valuable to provide a dynamic picture of such interactions during the infection process,” the authors write. “It is conceivable that the general strategy of HAPTIP can be applicable to many bacteria or virus, thus contributing to the discovery and understanding of host–pathogen interactions in multiple infection systems.”
Salmonella bacteria fend off a cell’s immune defenses by creating a pocket within the cell, called a Salmonella-containing vacuole, in which to hide. The bacteria hijacks and uses hundreds of the cell’s proteins to do so, making identification of those proteins key to thwarting the bacteria.
The HAPTIP method involves labeling the salmonella bacteria with a diazirine group, a chemical group that creates covalent bonds between Salmonella proteins and host cell proteins when an ultraviolet light is shined on the cell. A chemical probe enriches all the crosslinked proteins and isolates them from the other cell extracts. Scientists can then use mass spectrometry to identify the proteins.
One of the method’s strengths is that it can work at any point after salmonella has been introduced to the healthy cell. In their findings, the scientists tested the method at 15 minutes, one hour and six hours after salmonella infected a cell and identified more than 400 proteins interacting with the salmonella bacteria.
“You can design any time point based on when you choose to shine the UV light on the cells,” Tao said. “By looking at which proteins are interacting with the bacteria at those different times, we can determine the method the bacteria are using to hijack the cell, which will differ as time passes.”
Developing strategies to treat foodborne illnesses that stem from bacteria like salmonella and E. coli could have significant impact globally. The World Health Organization estimates there are 600 million global cases of foodborne illnesses each year resulting in 420,000 deaths.
The U.S. National Institutes of Health and National Science Foundation, as well as the Natural Science Foundation of China, funded this research.
Writer: Brian Wallheimer, bwallhei@purdue.edu
Source: W. Andy Tao, taow@purdue.edu
PHOTO: W. Andy Tao and colleagues have developed a method to implant a chemical label that acts like a GPS tracker into live salmonella bacteria. Once inside the bacteria, the probe can be captured at any given time, showing in real time the proteins interacting with the bacteria. (Photo courtesy W. Andy Tao).